The role of alternative poly-adenylation in gene regulation

The 3’ untranslated region (3’ UTR) of genes plays an important role in post-transcriptional regulation, affecting mRNA stability, localization and translation rate. The 3’ UTR length is determined by the site of poly-adenylation, Multiple sites can give rise to different 3’ UTR lengths (alternative poly-adenylation – APA), producing differentially regulated isoforms. In the lab, we use both bulk and single cell RNA-seq to determine the changes in APA during normal differentiation, or upon viral infection. Using our single cell data we are also studying the variability in isoform usage between cells in a uniform population - “biological noise”.
A single-cell based approach to study transcriptomic heterogeneity in prenatal clinical samples from congenital human cytomegalovirus (HCMV) infections
Human cytomegalovirus (HCMV) is the leading cause of congenital infection worldwide, affecting 0.5 to 2% of live births. Following HCMV infection, the host cells undergo major gene expression changes, and both viral and host genes have been shown to undergo extensive alternative splicing and APA, resulting in global changes in 3’UTR isoform expression. We are working to identify the changes in APA that occur during viral infection, and whether the severity of infection can be determined from this. In collaboration with Prof. Dana Wolf, we are analyzing the transcriptome of placental tissues infected in-vitro with HCMV. In placental samples taken from women that had been previously exposed to HCMV (seropositive), more changes are seen in gene expression than in seronegative samples, but interestingly the fewer changes in gene expression in the seronegative samples are accompanied but more changes in isoform usage. In the future, we plan to perform these experiments at the single cell level to identify the infected cells, and interactions between the different cells in this complex tissue, which acts as the “gate keeper”, protecting the embryo from infection.

APA changes in differentiation
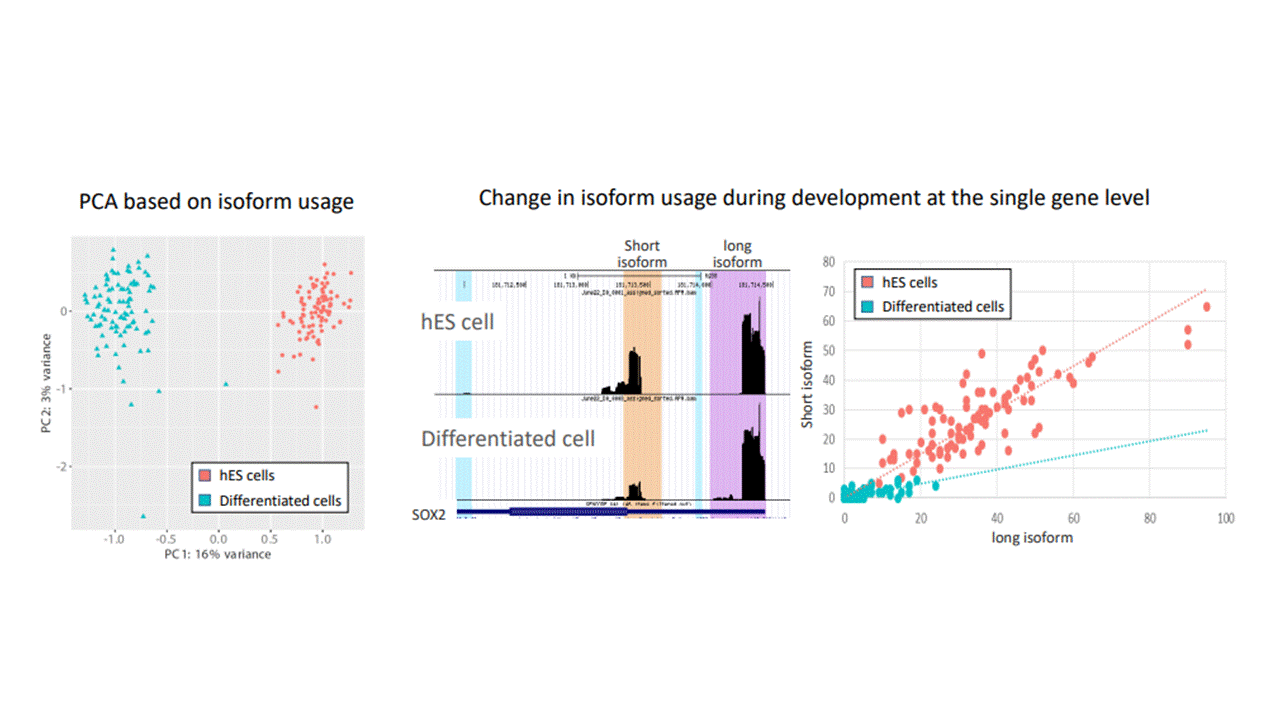
Differentiation of pluripotent embryonic stem cells to different cell types is achieved through gene regulation at its many levels. Here we use the CEL-Seq2 protocol to determine APA in single cells during spontaneous differentiation of human embryonic cells, and identify hundreds of genes in which the 3’ UTR isoform changes between the pluripotent and differentiated states. Isoform usage of genes expressed in both the pluripotent and differentiated cells is enough to distinguish cell identity. We are also using 10X technology for higher throughput experiments. Our aim is for specific genes to understand the factors causing the change in isoform usage, and the role of such changes.
3’ UTR isoform usage is highly consistent among single cells at the same cell state
RNA-seq analysis can determine the 3’ UTR isoforms present, and shows changes in isoform usage in different cell types. When identifying multiple isoforms of a specific gene in bulk samples, this could result from all cells having multiple isoforms, or each cell expressing a different isoform. Single-cell RNA-seq studies have attempted to determine APA in single cells. Due to low sensitivity of most protocols, cells are pooled according to cell type before looking at APA, thus preventing the question of cell-cell variability from being answered. When using the CEL-Seq2 protocol to determine APA in single cells, the sensitivity of the protocol and high sequencing depth allow us to determine the isoform usage at the single cell level without the need for pooling. To distinguish between biological changes in isoform usage and technical noise we also analyzed technical replicates of an RNA sample diluted down to single cell amounts. We show that the variability in isoform usage between cells is low, and very similar to that obtained for technical replicates. This is different from the overall gene expression that is higher in the cells. This suggests that 3’ end choice is tightly regulated at the single cell level, and is highly consistent among individual cells in homogenous populations.
